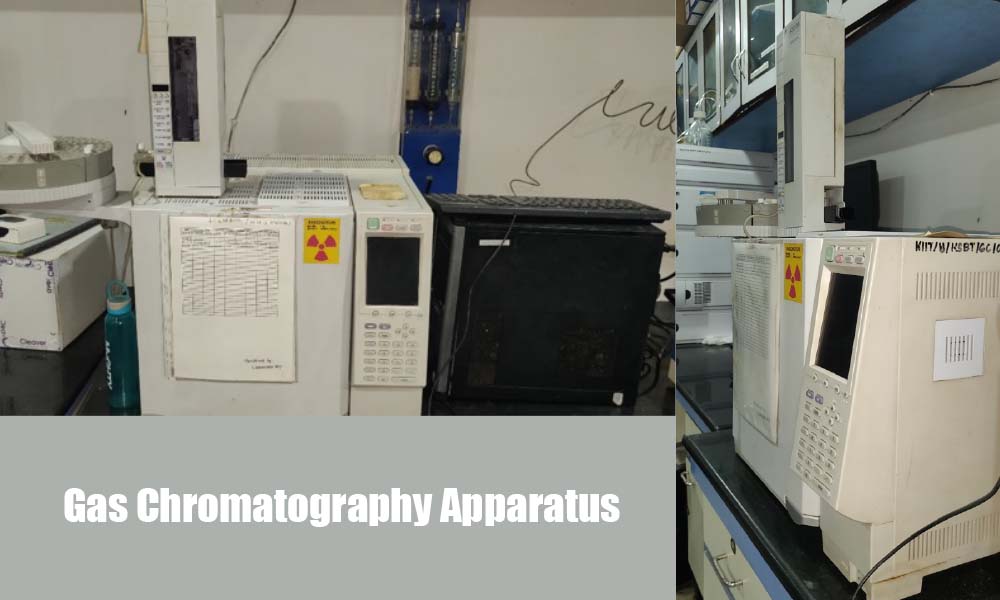
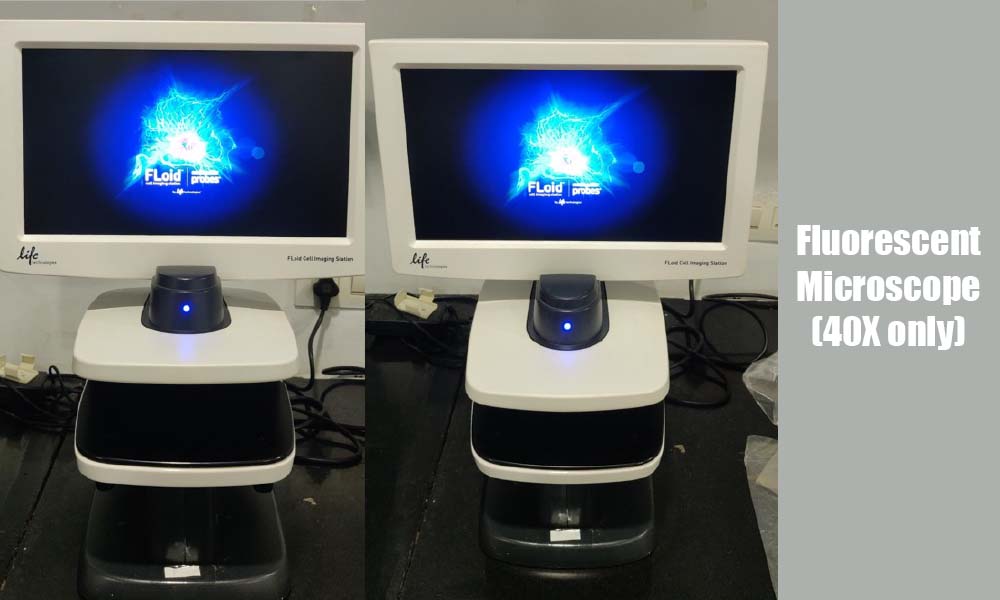
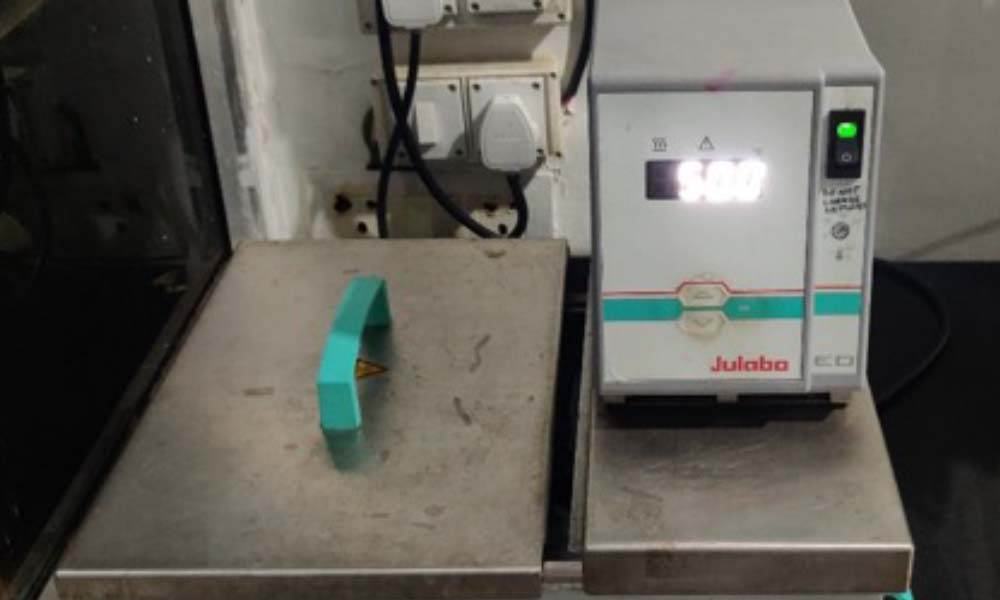
Laboratory
2-D PAGE, Protean i12 Apparatus
- Used for separation and fractionation of complex protein mixtures from tissues, cells, or other biological samples. It allows separation of hundreds to thousands of proteins in one gel.
- Applications of 2-DE include whole proteome analysis, cell differentiation, detection of biomarkers and disease markers, drug discovery, cancer research, bacterial pathogenesis, purity checks, microscale protein purification, and product characterization.
Dynamic Light Scattering (DLS) Apparatus
- DLS is most commonly used to analyze nanoparticles. Examples include determining nanogold size, protein size, latex size, and colloid size. In general, the technique is best used for submicron particles and can be used to measure particle with sizes less than a nanometer.